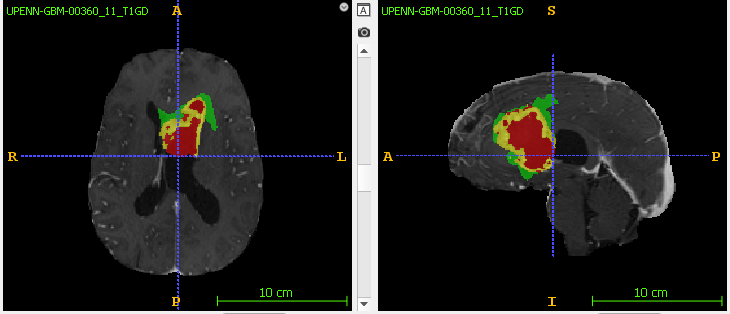
Introduction
The goal of brain tumor segmentation is to accurately detect and mark the location and extent of the tumor tissue from normal tissue so accurate surgical treatment can performed. Since Glioblastoma is fast growing, there may not be a clear demarcation between healthy and tumorous tissue. In addition, brain tumors have sub-regions such as enhancing tumor and edema (swelling) that will also need to be clearly segmented. 3D-MRI scans with multiple modalities (or parameters) are used to provide a spatial view that helps in accurate tumor segmentation. In this study, the authors have provided the following MRI sequences - T1 (spin lattice relaxation), T1-GD (also called T1 post contrast, T1 contrast enhanced), T2 (spin-spin relaxation), an T2-FLAIR (Fluid Attenuation Inversion Recovery).
How can we use AI - in this case Deep Learning - to segment brain tumor MRI scans? In this post, we will look at downloading and analyzing a Glioblastoma brain tumor dataset from TCIA. If you want a description of TCIA and other cancer related data portals, read my earlier post here. In future posts, I will walk through how to train and test a segmentation model from scratch. I use Google Drive and Colab for all my training, so the directions below pertain to these platforms only.
Step 1 - Download the data
I recommend downloading the NIfTI files as they are relatively easier to work with for a beginner. It is also smaller in size compared to the DICOM files. Finally, we want to work with 3D-MRI data, so let’s not use the histopathology images for this exercise.

Let’s first download the multiparametric MRI data. You’ll need to install the IBM Aspera Plugin in your browser to download the dataset. This plugin will allow you to pause and resume downloads if needed. When you click on the Download button, Aspera plugin will launch and ask to be added as a browser extension if you are using Chrome.
Once the Aspera extension is installed, click on the NiFTI-files folder to browse the package contents. We don’t need to download the entire package for this project, so let’s save some download time and space. Let’s see what we have in the package -
automated_segm - contains tumor segmentation files generated automatically by the study’s authors. Each patient has one “automated approximate segmentation” file. Later, we can compare our segmentation file with this dataset to see how our model fares.
images_DSC - contains MRI scans collected using the Dynamic Susceptibility Contrast (DSC) technique. Each patient has MRI scans for DSC, relative Peak Height (PH), Percent Signal Recovery (PSR), and relative Cerebral Blood Volume (rCBV). We will not use this dataset for our project.
images_DTI - contains MRI scans collected using the Diffusion Tensor Imaging (DTI) technique. Each patient has MRI scans that assess white matter (WM) sensitivity through different metrics such as Repetition Time (TR), Radial Diffusivity (RD), Fractional Anisotropy (FA) and Axial Diffusivity (AD). We will not use this dataset for our project.
images_segm - contains tumor segmentation files, one for each patient, manually segmented by expert radiologists. This will be our ground truth (GT) dataset that we will compare our AI generated segmentation files to.
images_structural - contains the skull stripped images. This will be our primary dataset. It has one sub-folder for each of the 630 patients that were part of the UPenn GBM study. Under each patient’s sub-folder, there are 4 NIfTI files (T1, T1GD, T2 and FLAIR), each corresponding to one MRI mode.
images_structural_unstripped - contains raw, skull unstripped MRI data, one sub-folder for each patient. We want to run our segmentation model training on skull stripped images, so we can ignore this folder.
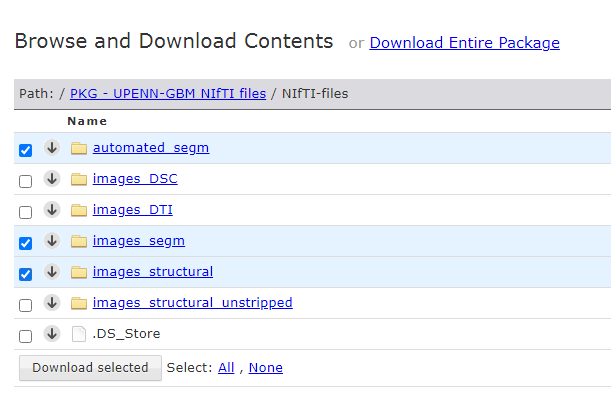
Go ahead and select these 3 folders and click the download button - automated_segm, images_segm, images_structural. The size is around 5.5 GB. Based on your internet speed, this will take a while. It took me several retries to finish downloading the files.

Don’t forget to go back to the main TCIA UPenn GBM collection page and download the clinical data file as well.
UPENN-GBM_clinical_info_v1.0.csv - Comma Separated Values (CSV) file with patient clinical information such as age, gender, genomics, extent of resection. We may potentially use this data later to see if we can conduct survival analysis.
Step 2 - Upload your data to Google drive
You can skip this step if you’re running the training on your local machine. The UPenn GBM NIfTI dataset is 69 GB in size. I first downloaded the data to my laptop, unzipped it, and then uploaded it to my Google Drive. In my example below, I created a folder called UPENN-GBM and uploaded the files there. You can create a folder with any name you like, but you will need to reference your particular file path in your code.

In our next post, we will analyze the dataset and determine our approach to automated segmentation using deep learning. Stay tuned.